
What's New
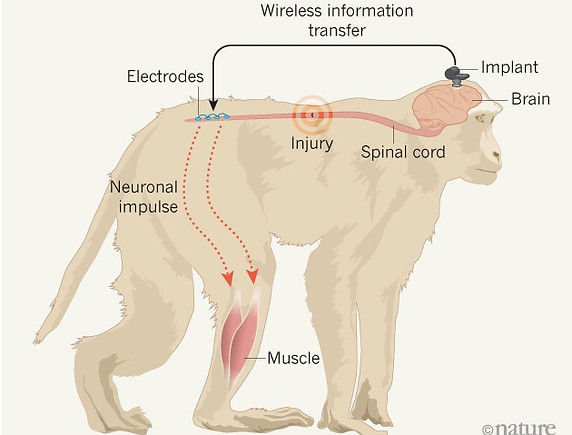
A wireless implant bypasses spinal-cord injuries in monkeys, enabling them to move their legs. (Adapted from A. Jackson Nature 539, 177–178 (2016))
For more than a decade, neuroscientist Grégoire Courtine has been flying every few months from his lab at the Swiss Federal Institute of Technology in Lausanne to another lab in Beijing, China, where he conducts research on monkeys with the aim of treating spinal-cord injuries.
The commute is exhausting — on occasion he has even flown to Beijing, done experiments, and returned the same night. But it is worth it, says Courtine, because working with monkeys in China is less burdened by regulation than it is in Europe and the United States. And this week, he and his team report1 the results of experiments in Beijing, in which a wireless brain implant — that stimulates electrodes in the leg by recreating signals recorded from the brain — has enabled monkeys with spinal-cord injuries to walk.
“They have demonstrated that the animals can regain not only coordinated but also weight-bearing function, which is important for locomotion. This is great work,” says Gaurav Sharma, a neuroscientist who has worked on restoring arm movement in paralysed patients, at the non-profit research organization Battelle Memorial Institute in Columbus, Ohio.
The treatment is a potential boon for immobile patients: Courtine has already started a trial in Switzerland, using a pared-down version of the technology in two people with spinal-cord injury.
“This study helps to open exciting new pathways to clinical studies and new bioelectronic treatment options for patients living with paralysis,” says bioengineer Chad Bouton, who researches medical devices used to bypass spinal-cord injuries at the Feinstein Institute for Medical Research in Manhasset, New York.
From rats to primates
The experiments are more of a progression than a sudden breakthrough: they are based on a decade of work in rats, Courtine says, and the monkeys reacted in very similar ways. The team first mapped how electric signals are sent from the brain to leg muscles in healthy monkeys, walking on a treadmill. They also examined the lower spine, where electric signals from the brain arrive before being transmitted to muscles in the legs. Then they recreated those signals in monkeys with severed spinal cords, focusing on particular key points in the lower part of the spine.

This is a 3D atomic scale map, or molecular structure, of the 2019-nCoV spike protein. The protein takes on two different shapes, called conformations—one before it infects a host cell, and another during infection. This structure represents the protein before it infects a cell, called the prefusion conformation. Credit: Jason McLellan/Univ. of Texas at Austin
AUSTIN, Texas — Researchers from The University of Texas at Austin and the National Institutes of Health have made a critical breakthrough toward developing a vaccine for the 2019 novel coronavirus by creating the first 3D atomic scale map of the part of the virus that attaches to and infects human cells.
Mapping this part, called the spike protein, is an essential step so researchers around the world can develop vaccines and antiviral drugs to combat the virus. The paper was published Wednesday, Feb. 19 in the journal Science.
The scientific team is also working on a related viable vaccine candidate stemming from the research.
Jason McLellan, associate professor at UT Austin who led the research, and his colleagues have spent many years studying other coronaviruses, including SARS-CoV and MERS-CoV. They had already developed methods for locking coronavirus spike proteins into a shape that made them easier to analyze and could effectively turn them into candidates for vaccines. This experience gave them an advantage over other research teams studying the novel virus.
“As soon as we knew this was a coronavirus, we felt we had to jump at it,” McLellan said, “because we could be one of the first ones to get this structure. We knew exactly what mutations to put into this, because we’ve already shown these mutations work for a bunch of other coronaviruses.”
The bulk of the research was carried out by the study’s co-first authors, Ph.D. student Daniel Wrapp and research associate Nianshuang Wang, both at UT Austin.
Just two weeks after receiving the genome sequence of the virus from Chinese researchers, the team had designed and produced samples of their stabilized spike protein. It took about 12 more days to reconstruct the 3D atomic scale map, called a
molecular structure, of the spike protein and submit a manuscript to Science, which expedited its peer review process. The many steps involved in this process would typically take months to accomplish.
